Bacteria Community Assessment
In a recent study, researchers found that microbial community diversity within streams in Southern California was related to pollution and surrounding land use (Ibekwe et al., 2016). Generally speaking, their results suggested that microbial community diversity decreases with
increasing pollution levels and urbanization (Ibekwe et al., 2016). If this trend is also true for the Milwaukee River Basin, microbial community diversity could be used as a novel parameter to help evaluate the water quality of streams and rivers.
Though previously a difficult parameter to measure, recent advances in technology have improved our ability to analyze microbial communities. This is largely due to the development of Next Generation Sequencing (NGS) technology, such as the Illumina MiSeq. Unlike traditional genetic approaches that focus on identifying a single bacteria group (such as qPCR), NGS technology can analyze a huge amount of DNA, allowing for the simultaneous identification of all major groups of bacteria within a single sample. The subsequent data can then be used to calculate microbial community diversity.
The goal of this study is to determine whether microbial community diversity in streams within the Milwaukee River Basin can be explained by water quality parameters and surrounding land use. This will help to assess whether this type of bacteria data could be used as an indicator of overall water quality in future environmental monitoring. Furthermore, we will also use this data to assess if any traditional water quality parameters help shape microbial communities in streams. Finally, this data will be used to detect spatial trends in bacterial groups, if any, with a specific focus on fecal-associated bacteria. Fecal bacteria could be an indicator of illicit discharges or failing infrastructure that could be addressed to improve stream water quality.
Project Objectives
Objective 1: Assess the functionality of using microbial community diversity as a water quality indicator.
1a). Is there a difference between bacterial community diversity in streams with “good” versus “impacted” water quality?
1b). Is microbial community diversity related to the surrounding land use at different stream sites?
Objective 2: Assess which traditional water quality parameters help shape the microbial community in streams.
2a). Which traditional water quality parameters correlate with microbial community diversity in streams?
Objective 3: Determine if spatial trends exist in fecal-associated bacteria throughout the Milwaukee River Basin.
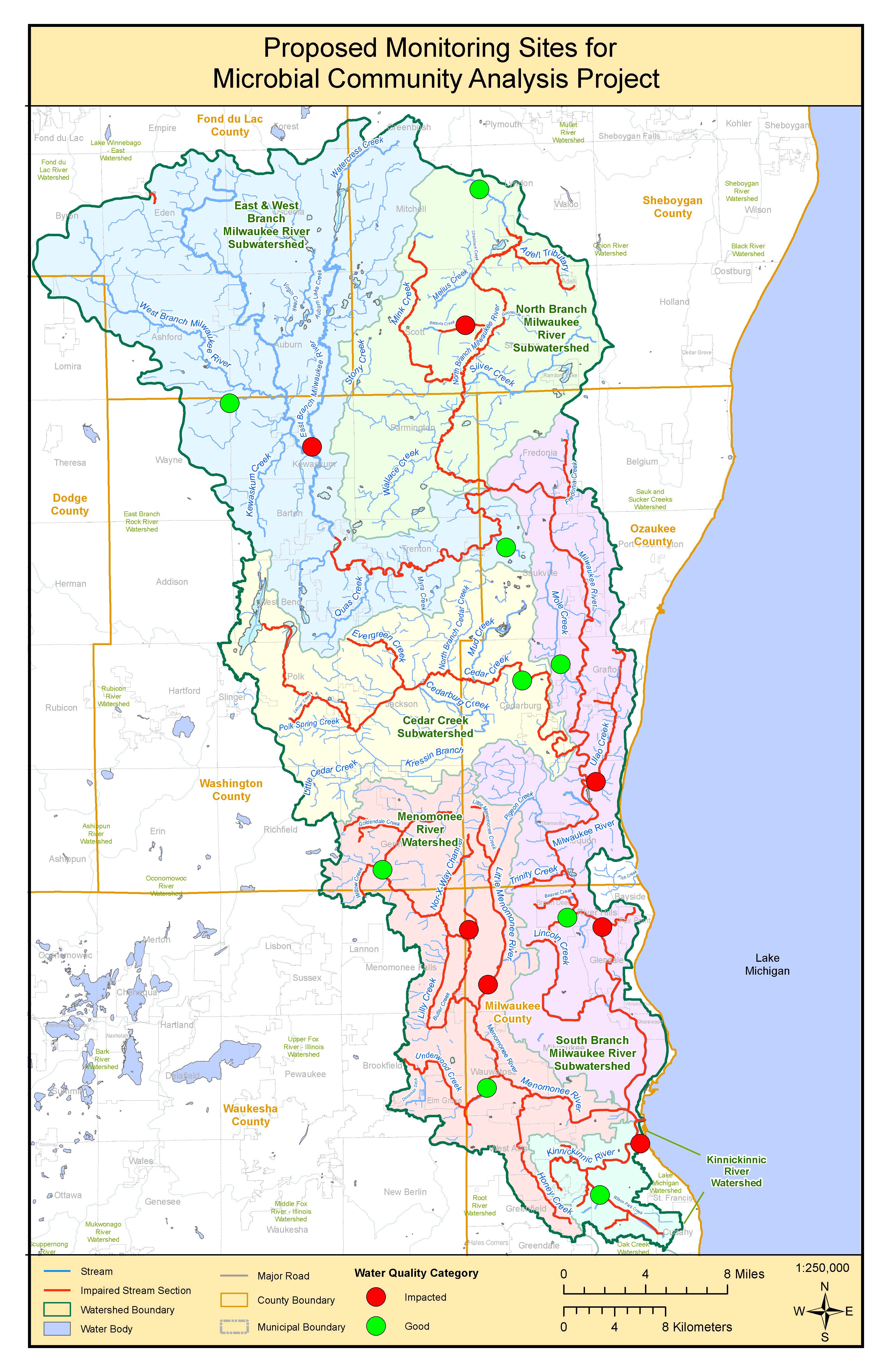
Site Selection
Sixteen sites were selected throughout the Milwaukee River Basin. All sixteen stations are relatively similar in size, favoring smaller tributary stations compared to the larger main river branches with larger water volume. To help answer question 1a., stations were defined as either “good” or “impacted” using all available dissolved oxygen (DO) data collected from the last ten years. Sites that exhibit chronically low DO over several years were defined as “impacted”, while sites with relatively high DO were defined as good. Eight of the total sixteen sites that were selected have “impacted” water quality while the remaining eight have “good” water quality.
The second consideration that was taken into account while selecting sites was their surrounding land use. In general, the Milwaukee River Basin has a south to north gradient of urban to rural land use. Eight of the total sixteen stations selected are within the southern urban half of the basin, while the remaining eight are found within the northern rural half of the basin.

Thanks to our amazing project partners!

Thanks to our funders for their generous support of this program!
